Tweet
March 31, 2016
Proceedings of the Natural Institute of Science | Volume 3 | SOFD 1
Laboratory "evolution" of complex life
M. Reysaim1 & K.M. Knoll2,3
1 - Whereabouts unknown
2 - Department of Science, Northern Connecticut State University (NoCoStU, "You get what you pay for"), USA
3 - Corresponding Author: profknoll57 at gmail.com
Submitted: March 13, 2016
Accepted: March 20, 2016

Introduction
Laboratory evolution of bacteria allows the natural creation of genetic variants to observe the course of natural selection. Evolution in the lab allows the control of selective conditions by the investigator as compared to the uncontrolled nature of selection in the wild. This report uses investigator-defined conditions to allow evolution (or whatever process might occur) to create complex cellular life beginning with a single cell of the bacterium Escherichia coli.
Methods
A 10 L Fernbach flask containing 1 L of dilute growth medium was gently agitated while incubating at 25°C (Figure 1). A sterile, 500 g flat stone with an attached, sterile castle (thus forming a rocky beach) was also placed in the flask to provide a more balanced and harmonious ecosystem (sensu, WWF). The flask was inoculated with a single cell of E. coli K12 from a frozen stock culture.
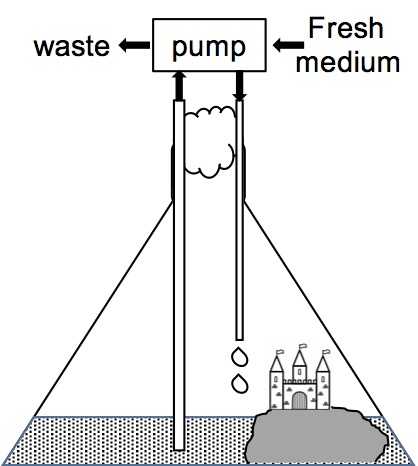
The cells doubled every 24 hours so that, starting with 1 cell on day 1 (0 generations, 20 cells), one week later there were 26 cells (6 generations, 64 cells). After 1 year there were 2364 cells (364 generations, 3.8 billion cells) at which time spent medium was continuously removed while an equal volume of fresh medium was added to the culture to maintain a constant volume, a constant cell density, and to provide fresh nutrients (Figure 1). Samples were removed at intervals and examined under a microscope to observe changes in the population, until, in some cases, the use of a microscope became unnecessary and a magnifying glass or simply the naked eye was used. To test the reproducibility of the method, this set-up was repeated for 10 independent cultures, each starting with a single cell from the same E. coli K12 frozen stock culture.
Results and Discussion
After 5,000 generations of E. coli and 3 generations of graduate students (13.7 years), the 10 independent cultures were examined microscopically and macroscopically (Table 1). Seven of the cultures contained only E. coli cells. The genomes of cells harvested from each of these 7 cultures were sequenced and each cell had, as expected, an average of 16.7 mutations in a total 5 million nucleotide pair genome (Figure 2).
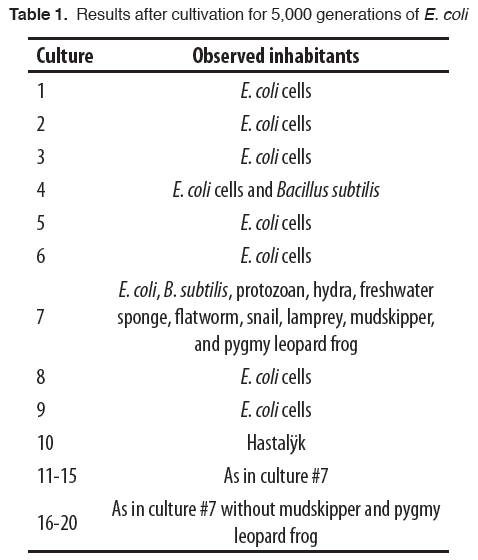
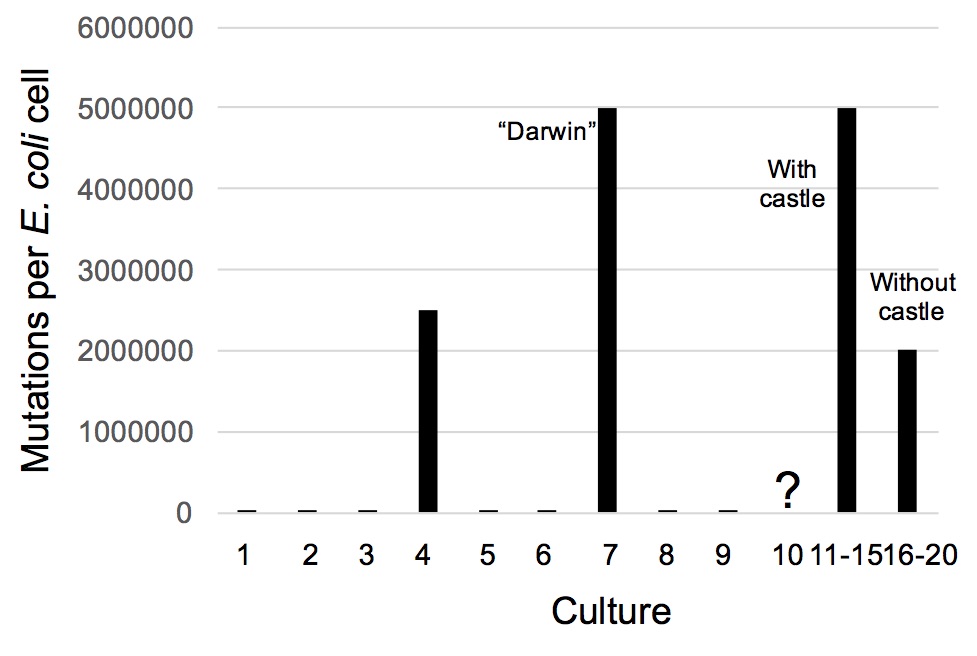
Culture #4 contained half E. coli and half unusually large cells. Based on their genome sequences, these larger cells were Bacillus subtilis, a bacterium distantly related to E. coli. All of its nucleotides were different than those of the starting E. coli indicating that 2.5 million mutations had occurred per cell (Figure 2).
Two explanations are offered for this surprising and unprecedented development. First, the culture may have been contaminated. The genome sequence of this B. subtilis was unlike those of Bacillus cells isolated from the laboratory (surfaces of countertops, equipment and graduate students) or of any Bacillus species ever discovered, so this possibility was ruled out. Second, this B. subtilis may have evolved from the E. coli. Although seemingly a ground-breaking discovery, it presents no violation of the biological principles outlined by the Discovery Institute (“Where we teach the controversy!”). The formation of one bacterium from another is simply the generation of a kind from the same kind and, according to the principles of baraminology, not the formation of one species from another (i.e., “evolution”; Gishlick 2006). Also, E. coli and B. subtilis have different genus and species names only because of an artificial naming convention that makes distinctions that do not even exist in nature (Doolittle 2012).
Culture #7 contained E. coli, B. subtilis, Paramecium aurelia (a protozoan), Hydra magnipapillata (a hydra), Spongilla lacustris (a freshwater sponge), Planaria torva (a flatworm), Lymnaea stagnalis (a snail), Lampetra aepyptera (a lamprey), Periophthalmus gracilis (the mudskipper, an air-breathing fish), and a pygmy leopard frog (Rana pipiens). The frog was sitting on the castle and the mudskipper was on the rock “beach.” The three vertebrates (lamprey, mudskipper, and frog) were all noticeably smaller than their counterparts in natural populations. This size discrepancy may have been due to a number of factors including: size restrictions of a 10 L flask, less available food resources, or the lab shaker causing individuals to bounce off the flask walls and suffer tissue damage.
Genome sequencing confirmed the visual identifications (data not shown). Since nearly all the E. coli cells had mutated, 5 million mutations had occurred per cell (Figure 2). Again, contamination of the culture was ruled out by genome sequencing of samples taken from surfaces and deep recesses in the laboratory (including those of the graduate students).
Since the chances of such a result seemed exceedingly rare, we set out to ascertain whether this result was a fluke. E. coli cells from culture #7 (now called strain “Darwin”) were inoculated into 10 new cultures, but only 5 of these (cultures #11-15) were provided with sterile castles/rocks. After 5,000 E. coli generations (3 more graduate student generations), the cultures were examined and the results are shown in Table 1 and Figure 2. Cultures #11-15 had the same spectrum of organisms as in culture #7. Cultures #16-20 had all but the mudskipper (P. gracilis) and the pygmy leopard frog (R. pipiens). Cultures #16-20 lacked a castle and rock so apparently the lack of dry land affording a place to breathe hindered their survival.
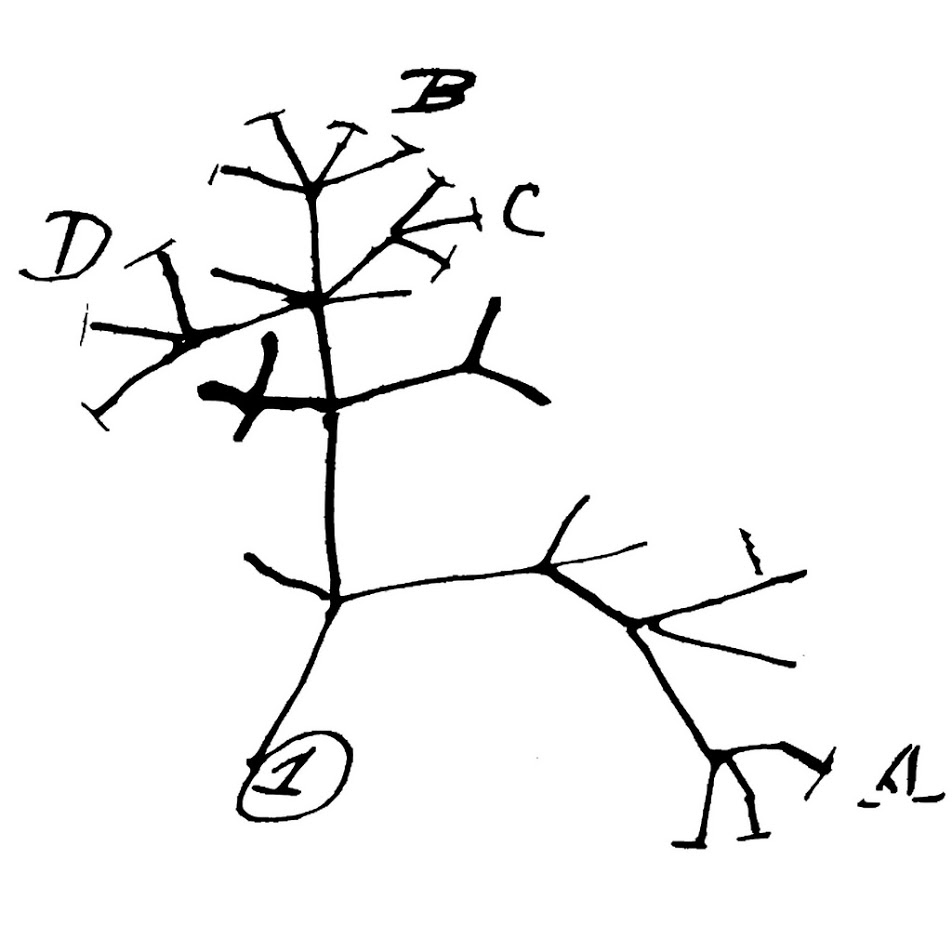
Genome sequencing of the Darwin strain showed that it had developed a highly defective DNA replication capacity. While making a new copy of its chromosome during cell division, the Darwin cells were extraordinarily sloppy in synthesizing DNA. This resulted in changing every nucleotide of DNA in every cell in the culture. The new, grossly mutated DNA was, however, misread by an illiterate DNA-reading machinery so that fully viable cells resulted. So instead of creating two new E. coli cells, cell division produced an E. coli cell and a cell of B. subtilis. The B. subtilis cells, having the same defective cellular machinery as their E. coli parents, sometimes gave rise to a protozoan (P. aurelia), which later gave rise to a fertilized egg of a freshwater sponge (S. lacustris). Those highly error-prone progeny cells then formed new kinds of animals, each more advanced than the previous. The relationships among the creatures are shown in Figure 3.
Culture #10 contained a single cell of the little-studied Hastalÿk (Lovecraft, H. P., personal communication) surrounded by the empty husks of billions of E. coli cells. Small amounts of DNA released by Hastalÿk were harvested from the cold, dark culture supernatant. DNA sequencing revealed that the genome of Hastalÿk uses all 26 letters of the English alphabet and some cryptic, ancient runes. Before seizing up and disintegrating into a fine black dust, the DNA sequencer produced data that could be read as an English document. It described an impending annihilation of humans, warning of an indescribable horror that would creep “like a stealthy wraith through the souls of weak-fleshed men” (see Supplemental Figure 1).
The Hastalÿk (Figure 4) apparently appeared in the 5,000th generation as the laboratory notes of graduate student M. Reysaim note the 4,999th generation population was only E. coli cells. M. Reysaim’s notebook is all that remains of M. Reysaim, as culture #10 was found unattended, M. Reysaim’s iPhone account was deleted, and his friends, family and landlord do not recall he ever existed. Room 324b, where culture #10 was incubated, has since been secured and is currently under the jurisdiction of NoCoStU (“No higher education!”) Chemical, Biological and Ecclesiastical Safety. We must never speak of this experiment again.

More Articles Below!

Proceedings of the Natural Institute of Science (PNIS) by https://instsci.org/ is licensed under a Creative Commons Attribution-ShareAlike 4.0 International License.